View Of A Streamlined Crispr Cas9 Approach For Fast Genome Editing In Toxoplasma Gondii And Besnoitia Besnoiti Journal Of Biological Methods
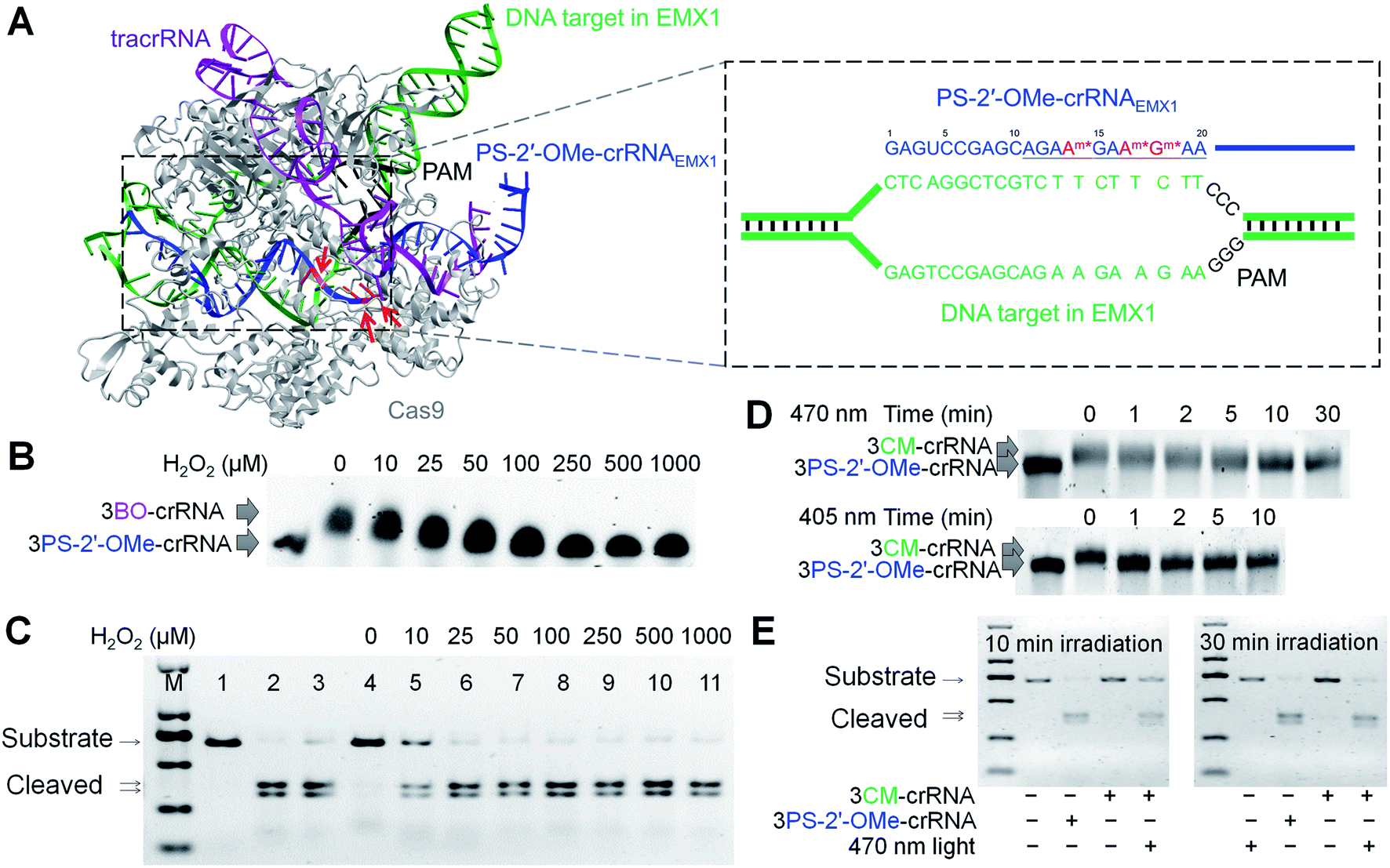
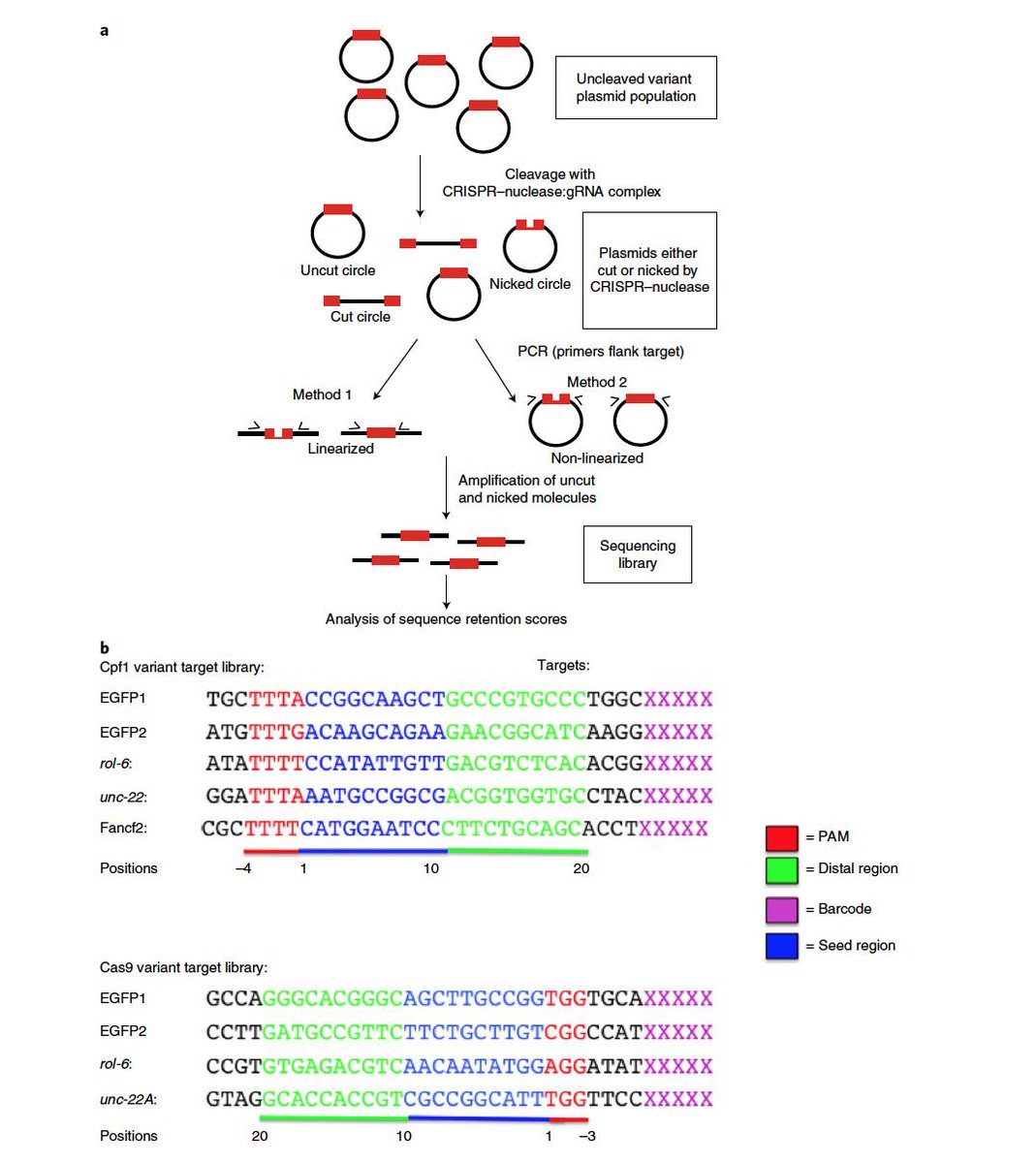
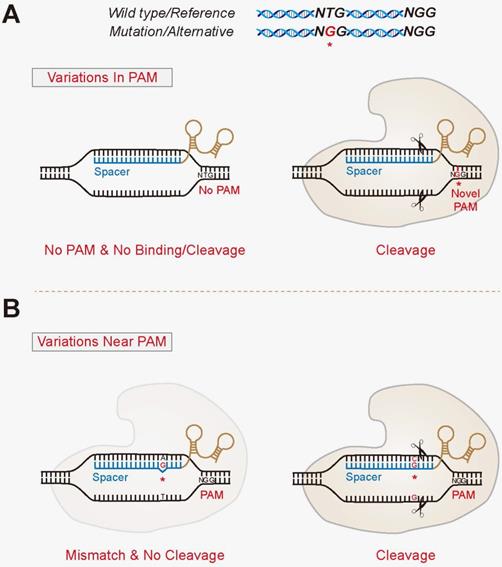
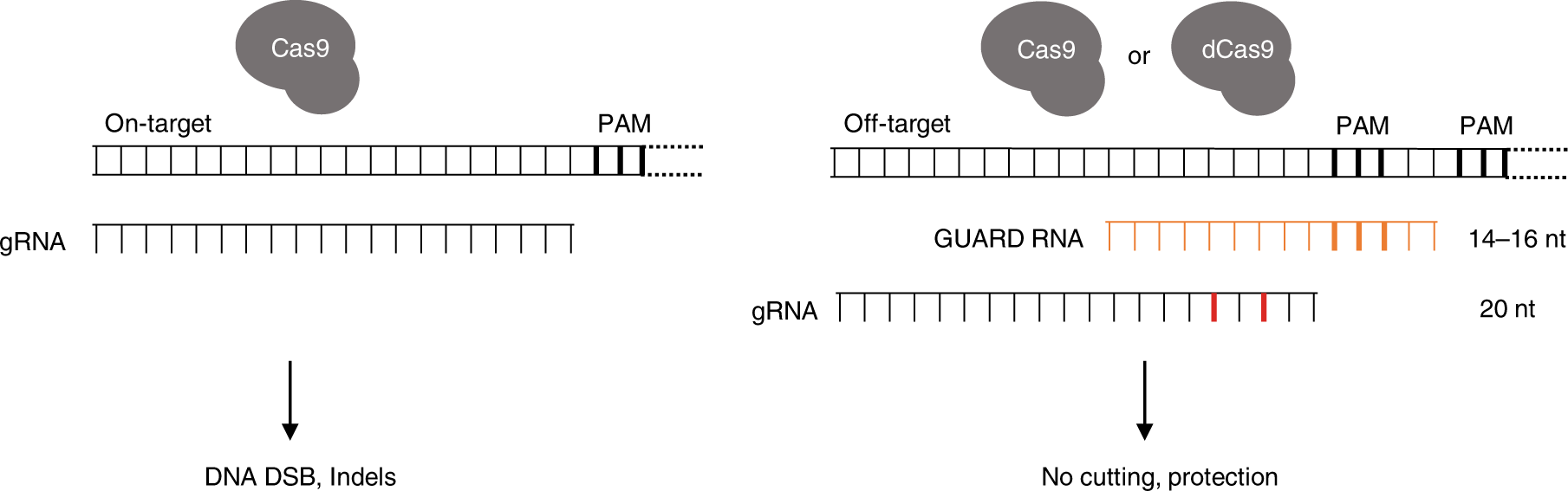
PAM (seed region) are especially important for target site recognition In many cases, offtarget mutations happen at sites where the gRNA seed region has no mismatches but the nonseed region does 3, 15 To knock out a specific region in the genome, such offtarget effects should be avoided Reducing such risk is espe cially important when we consider further application ofGRNA targeting region of interest gRNA targeting gene promoter elements gRNA targeting gene promoter elements Table 2 Plasmid Vector Expression System Components Applica on Constitutive or inducible Cas9 Constitutive or inducible gRNA Reporter/selection marker Expression of Cas9 and gRNA 3 wwwgenscriptcom Modified recombinant AAV particles are attractive for transduction
Grna seed region
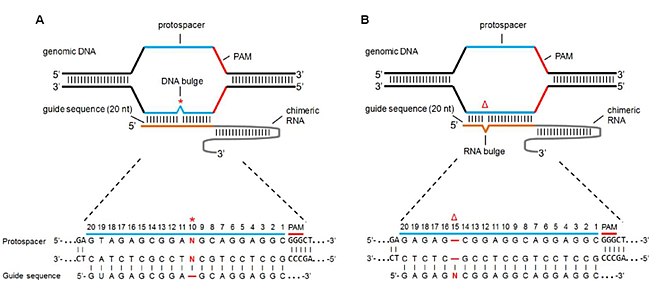
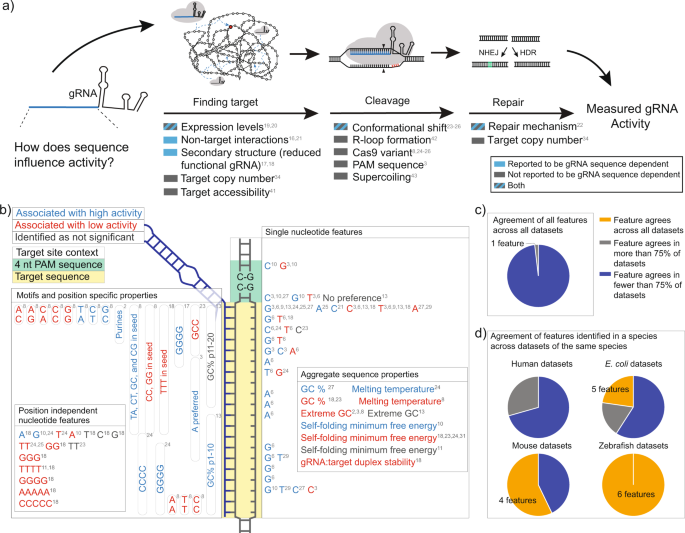
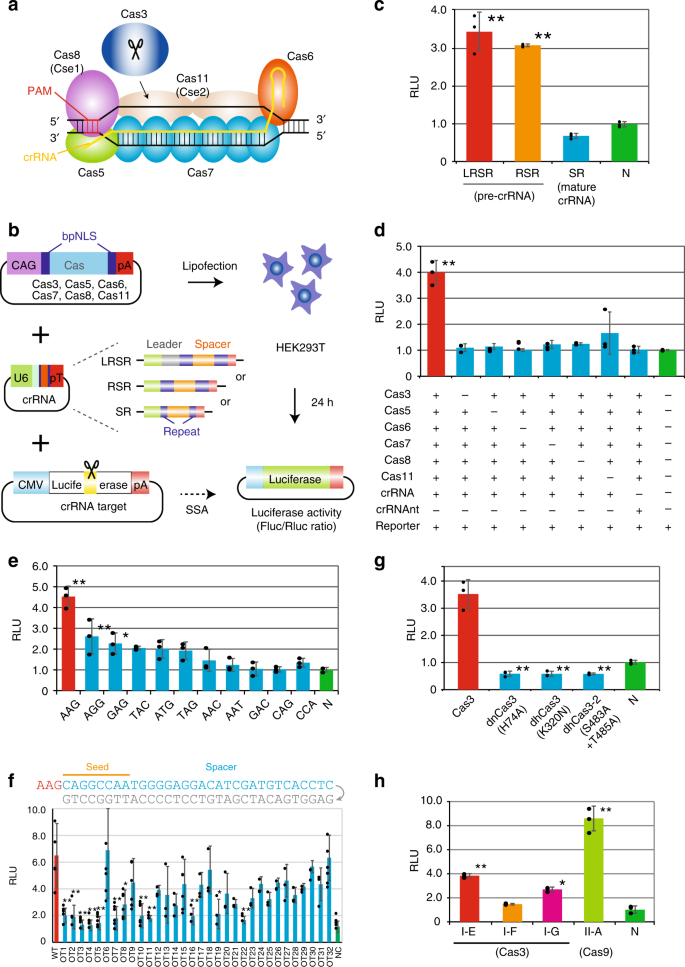
Grna seed region-The gRNA is a short synthetic RNA composed of a scaffold sequence necessary for Casbinding and a userdefined ∼ nucleotide spacer that defines the genomic target to be modified Thus, one can change the genomic target of the Cas protein by simply changing theMatches within the gRNA and reported varying degrees of specificity within the bases that constitute the spacer sequence Consistent with previous studies, they reported that mismatches within the PAMdistal region are more tolerated and defined a range of 8 to 14 bases in the PAMproximal region as the seed sequence that provides
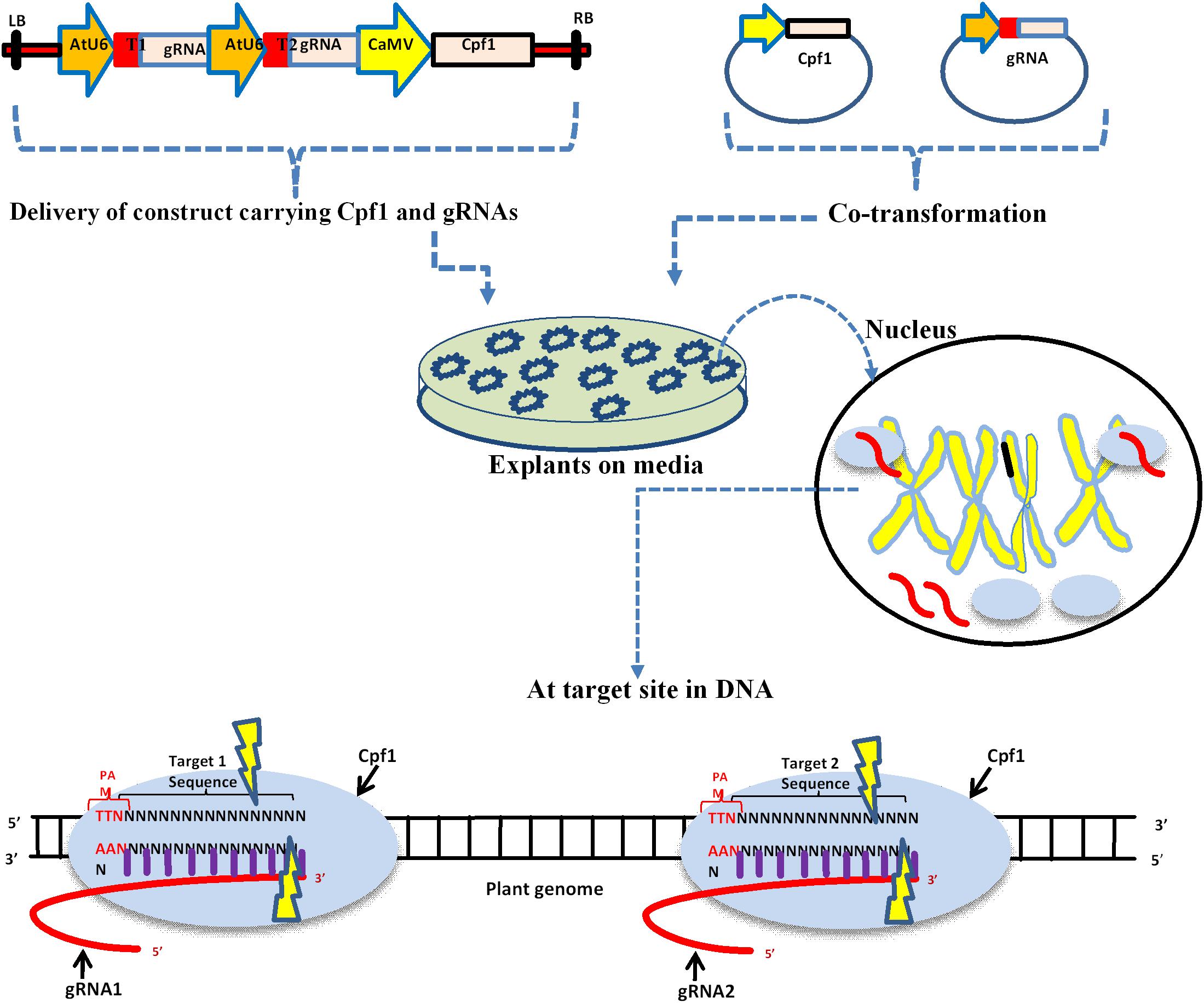
Rna Guided Genome Editing In Plants Using A Crispr Cas System Ppt Download
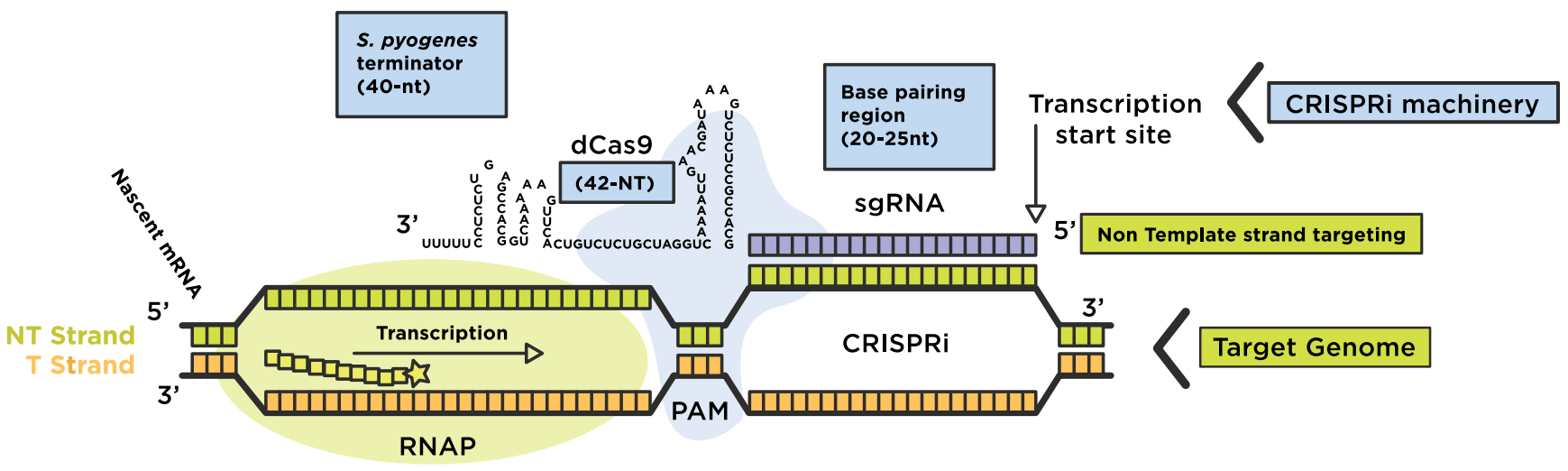
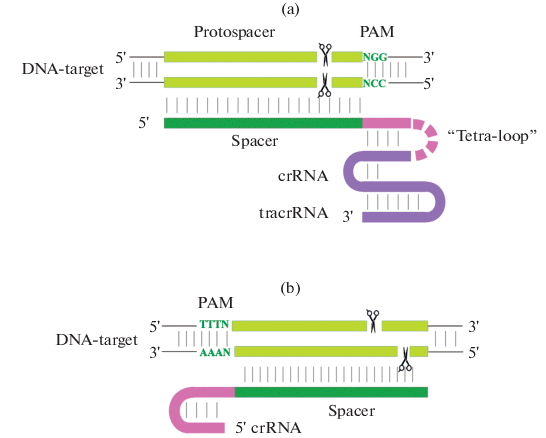
Once the PAM is recognized, the guide region of the gRNA undergoes seed nucleation to form an Aformlike helical RNADNA hybrid duplexGrna seed region Grna seed regionAn approach called CREATE enables multiplex genome engineering, protein engineering and mapping of mutations in bacterial and yeast cells Improvements in DNA synthesisDe Strasbourg à l'Est à NogentsurSeine à l'Ouest, la Région Grand Est s'étend sur 57 500 km² Elle compte 9 départements Ardennes, Aube, Collectivité européenne Of note, both motifs are located in the sgRNA "seed" region (the PAMproximal 10–12 bases of the targeting sequence) that is important for pairing with the target DNA (
Once the PAM is recognized, the guide region of the gRNA undergoes seed nucleation to form an Aformlike helical RNADNA hybrid duplex Only once the RNA and DNA complete Rloop formation, also known as the zipped conformation, and structural rearrangement of the nuclease domains commence, can the endonuclease cut the DNA creating a DSB (Jiang et301 Moved Permanently nginxThe seed sequence is essential for the binding of the miRNA to the mRNA The seed sequence or seed region is a conserved heptametrical sequence which is mostly situated at positions 27 from the miRNA 5´end Even though base pairing of miRNA and its target mRNA does not match perfect, the "seed sequence" has to be perfectly complementary
Grna seed regionのギャラリー
各画像をクリックすると、ダウンロードまたは拡大表示できます
 |  | |
 |  | |
 |  | |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  | |
 | ||
 | ||
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  | |
 |  | |
 |  | |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  |  |
 | ||
 |  | |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  |  |
 |  | |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  |  |
 |  |  |
 |  |  |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  |  |
 | ||
 |  |  |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  |  |
 |  | |
 |  | |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  | |
 |  | |
 |  |  |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 | ||
 |  |  |
 |  | |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  | |
 |  |  |
「Grna seed region」の画像ギャラリー、詳細は各画像をクリックしてください。
 |  | |
 |  |
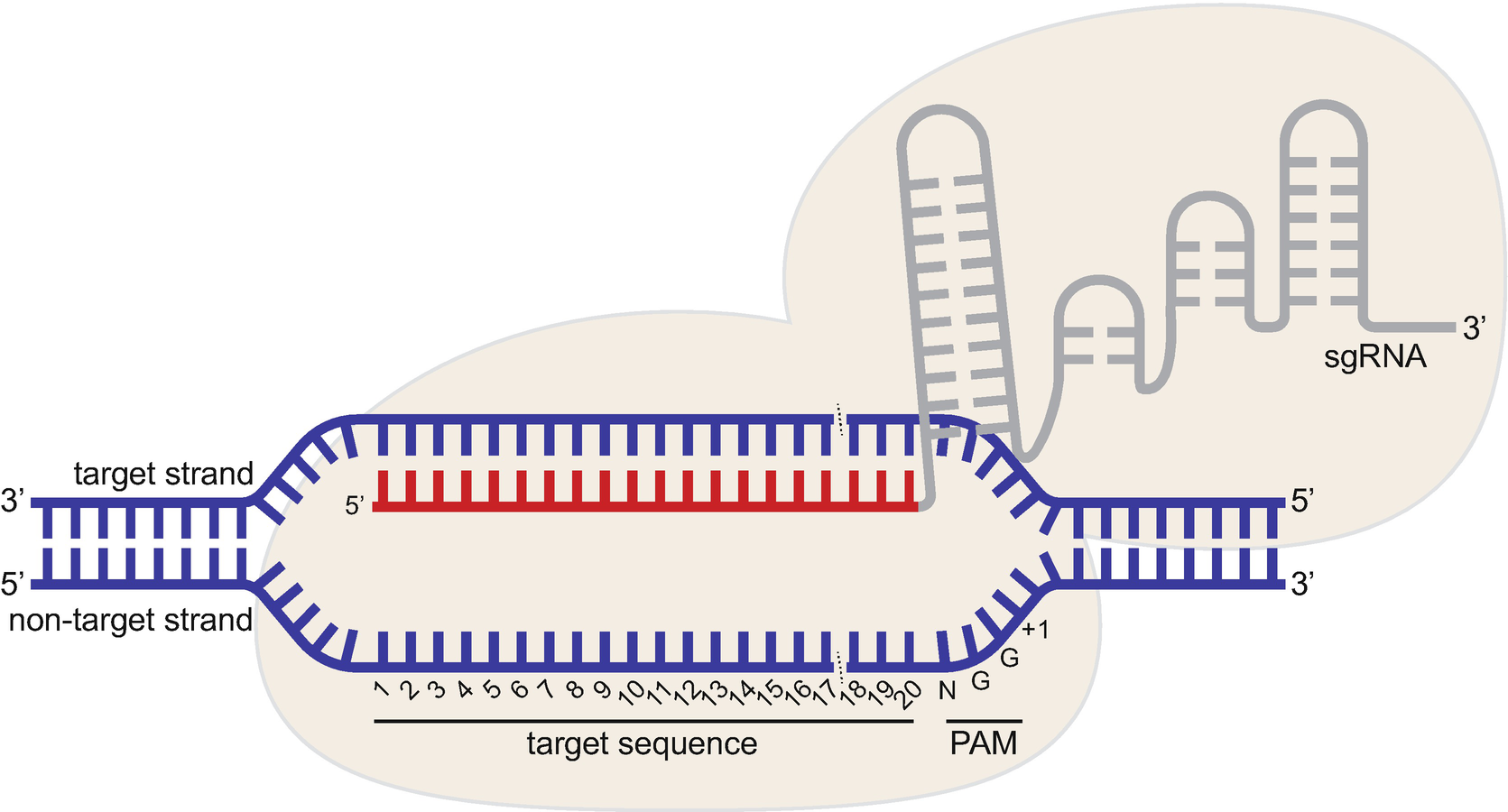
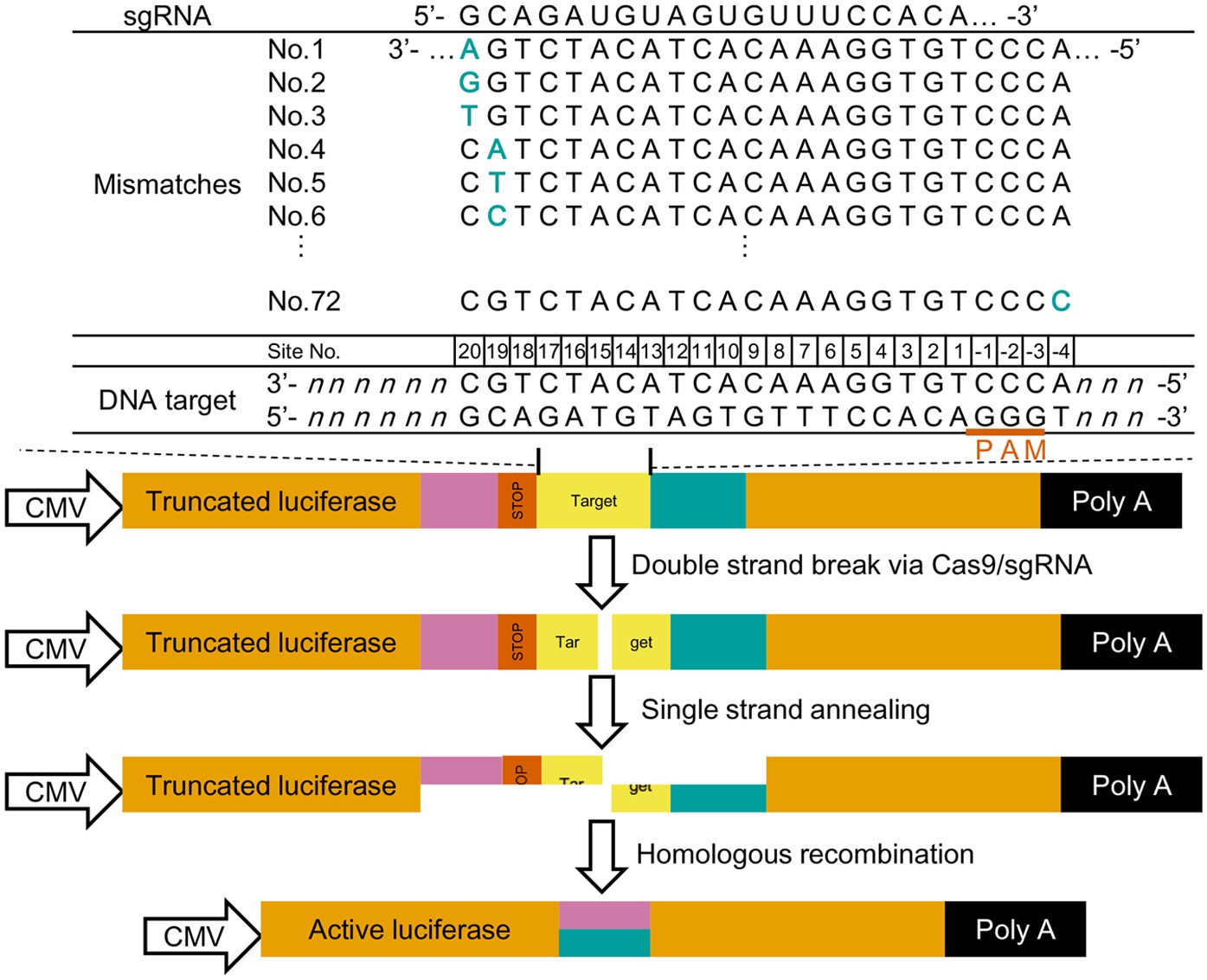
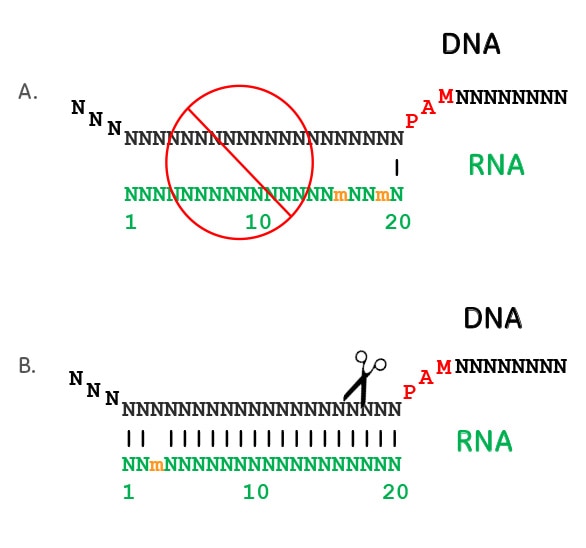
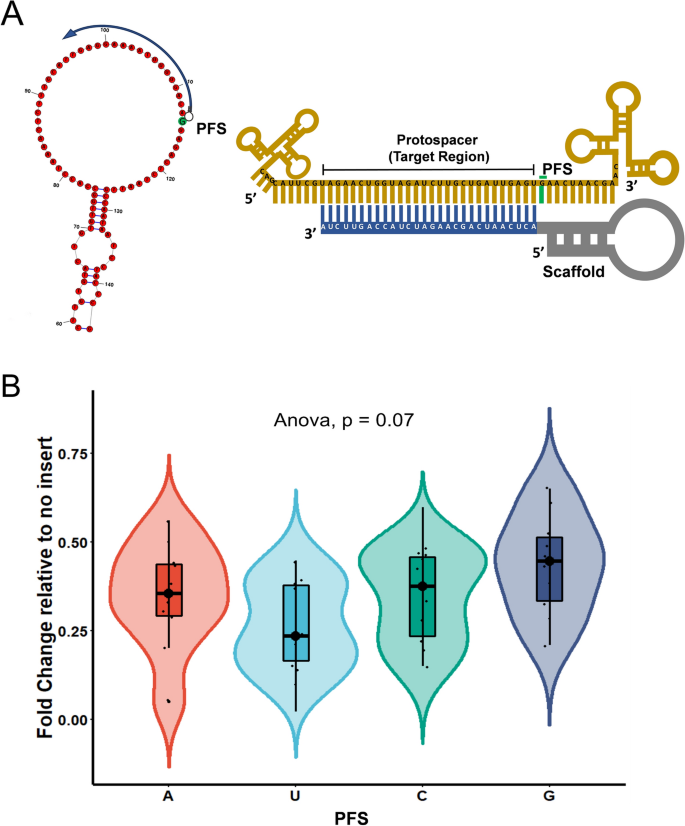
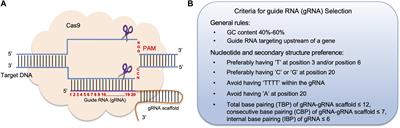
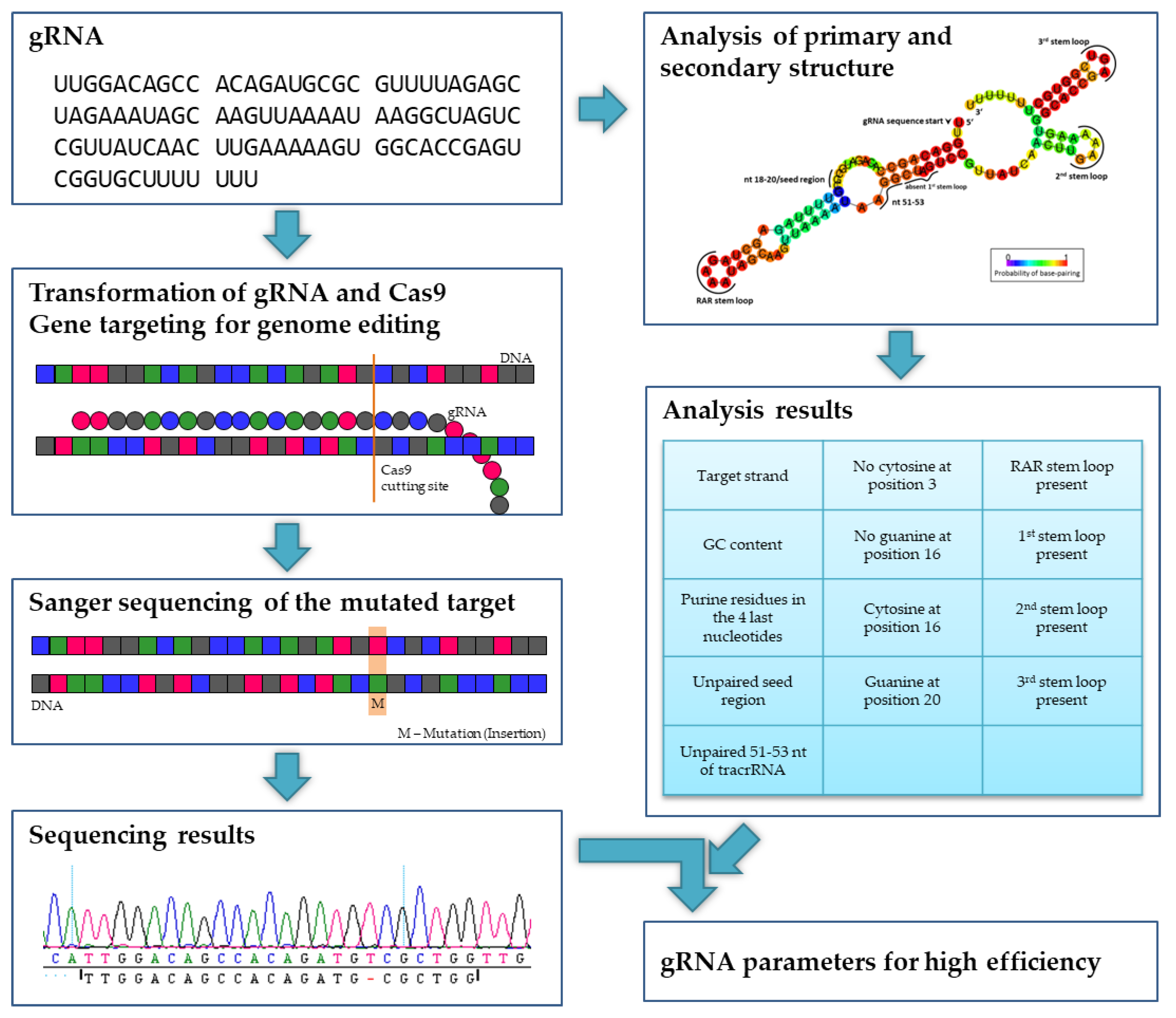
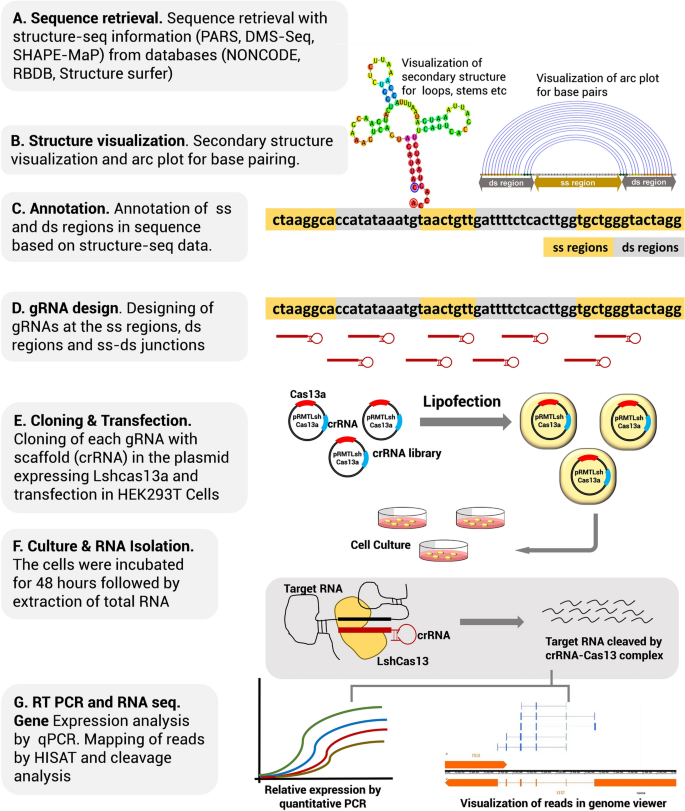
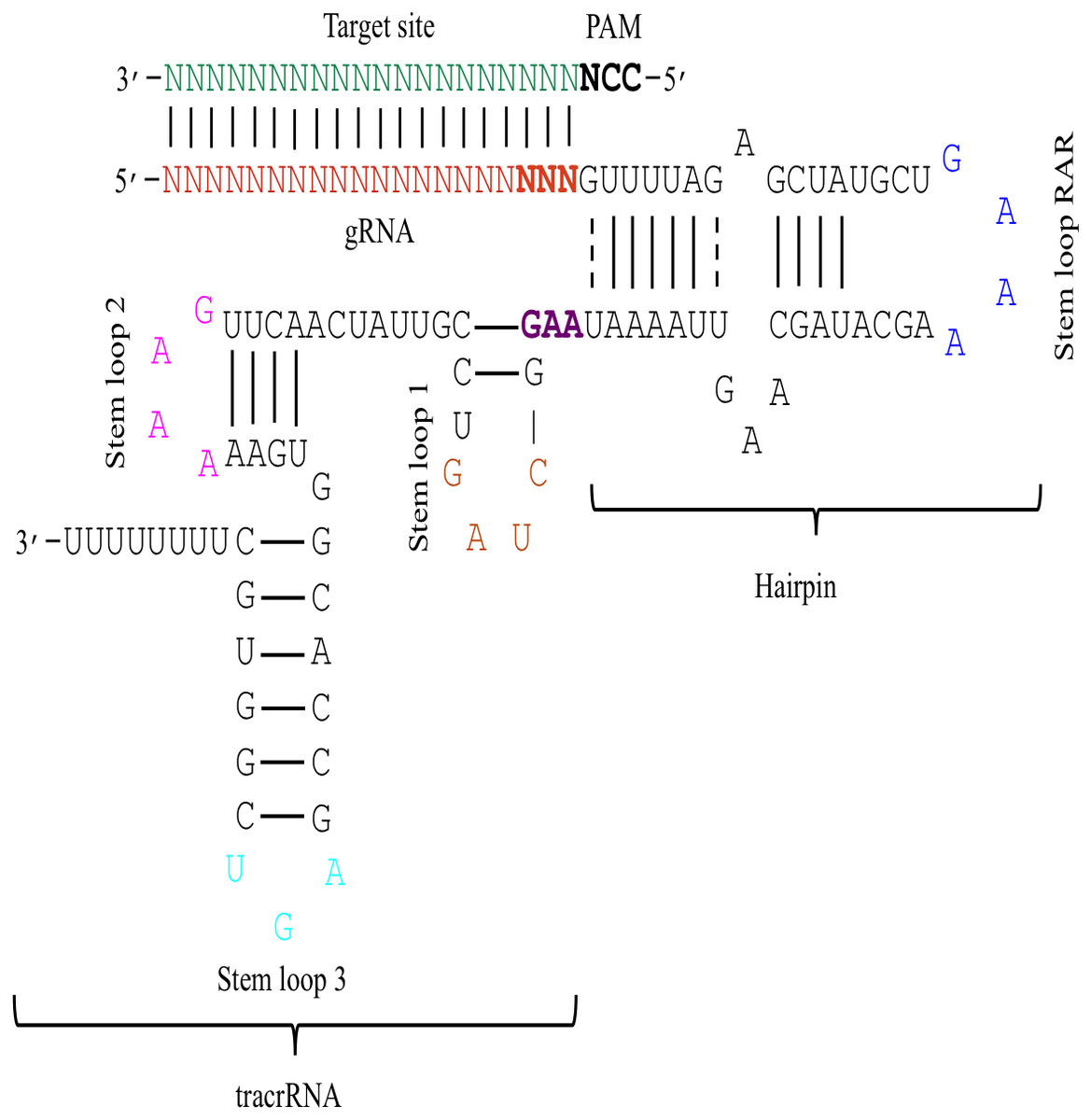
The 5′end of gRNA (gRNA seed) is shown to pair with one strand of targeted DNA The scaffold of gRNA is labeled with darkred cycles A PAM motif (NGG) is located adjacent to the DNA–gRNA pairing region in the complementary strand of targeted DNA The DNA–gRNA basepairing should be at least 15bp long Singlenucleotide mismatch in the gRNA seed region lowers frequency but does not prevent Cas9 cleavage It is well known that CRISPR/Cas9 may result in offtargeting at unintended sites which share sequence homology with the target site (Fu et al 13, Hsu et al 13) In this study, we used two gRNAs for simultaneous targeting of F3H The second gRNA (ie, gRNA3)
Incoming Term: grna seed region,




0 件のコメント:
コメントを投稿